Case History
A 59 year old African American female presented to the emergency department with fevers, weakness, fatigue and altered mental status. Her past medical history was significant for hypertension, diabetes mellitus (type 2) with end stage renal disease and a recent cerebrovascular accident the month prior. Her surgical history included a mitral valve repair surgery three years ago and a renal transplant two years ago. Current medications included prednisone, mycophenolate and tacrolimus immunosuppressive agents. Physical examination was unremarkable except for a temperature of 101°F and she was oriented to person, place and time. Pertinent labs included a WBC count of 13.2 TH/cm2, microcytic anemia, and a creatinine of 1.51 mg/dL. Due to previous cardiac surgery, a transesophageal echocardiograph (TEE) was performed and showed a large vegetation (1.6 x 1.5 cm) on the mitral valve. A diagnosis of endocarditis was made and the patient was started on broad-spectrum antibiotics & taken to surgery for a mitral valve replacement. Multiple blood cultures were negative to this point. Portions of the mitral valve were submitted to surgical pathology and the microbiology laboratory for bacterial, fungal and AFB cultures.
Laboratory identification
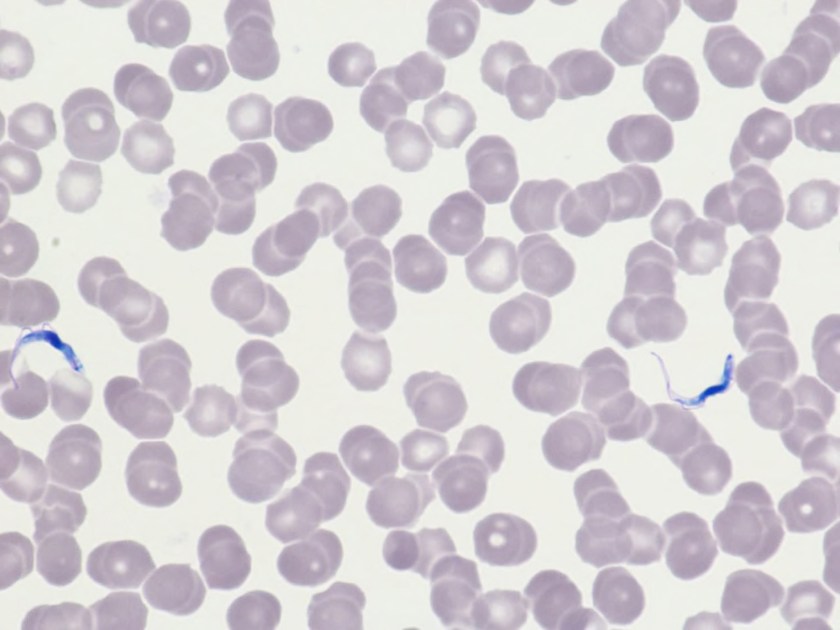
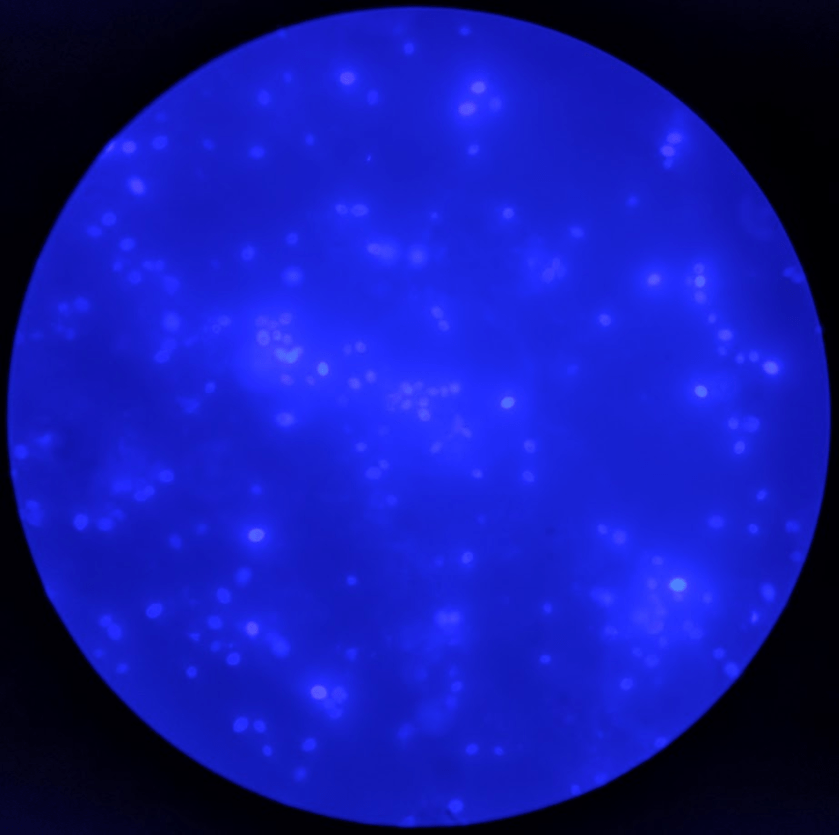
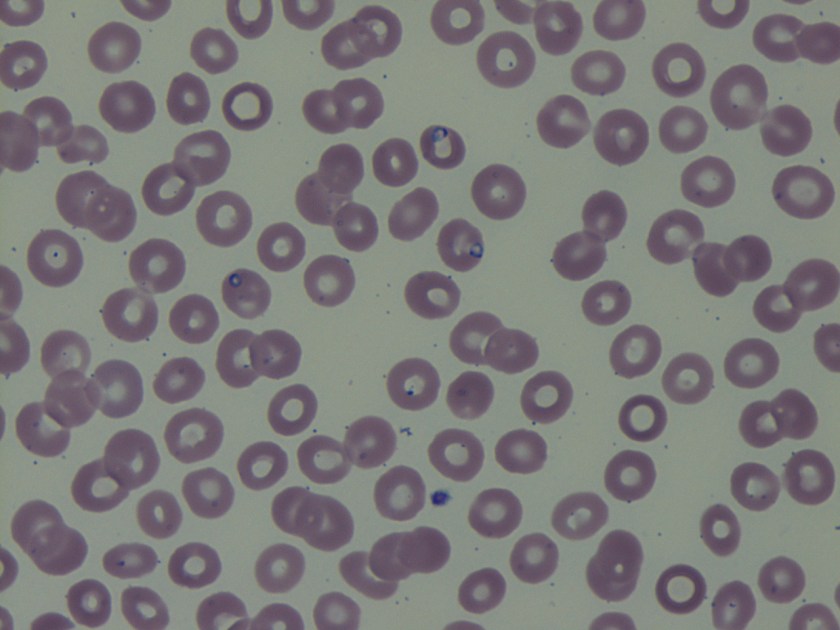
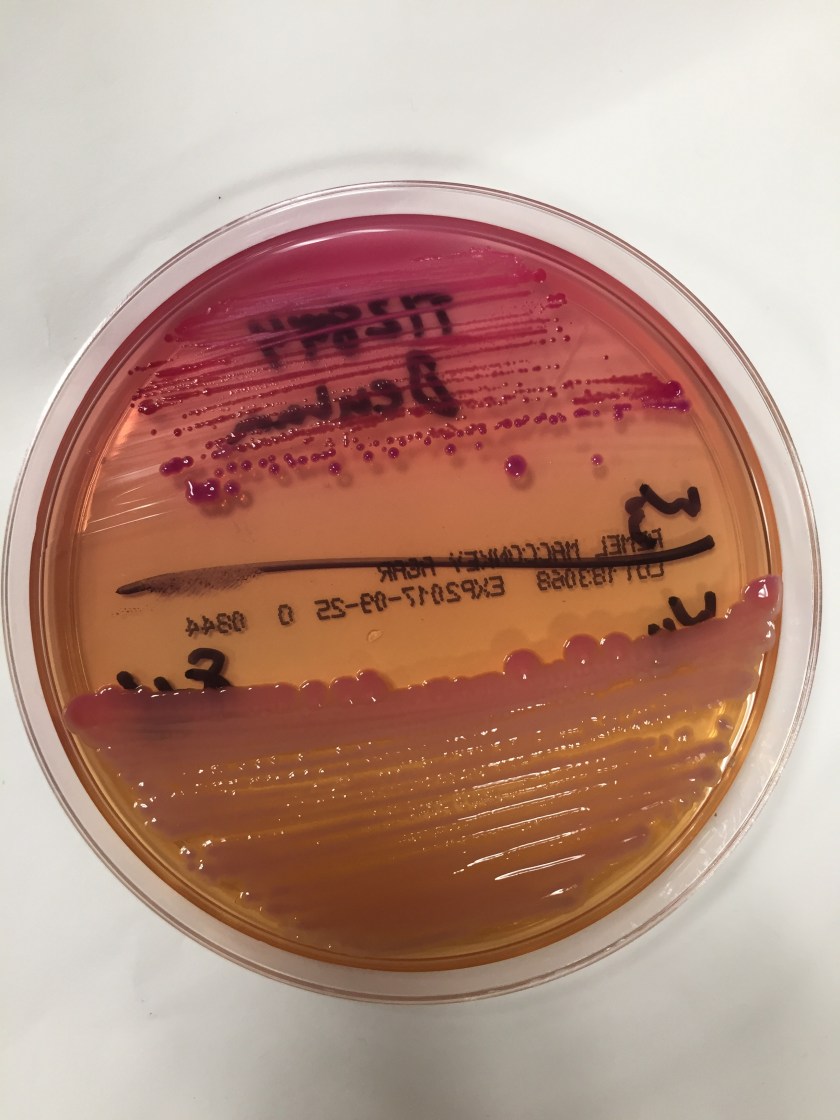
Surgical pathology received an aggregate of tan-yellow, fibrous tissue fragments (3.1 x 1.5 x 1.1 cm). Histologic assessment showed a confluent mass containing abundant narrow, septate hyphae consistent with a fungal infection (Image 1). No definitive pigment was identified. Grocott’s methenamine silver (GMS) stain also highlighted the narrow hyphae with numerous septations (Image 2). In the microbiology laboratory, a darkly pigmented mold grew after 5 days of incubation on Sabouraud dextrose agar (Image 3). Lactophenol cotton blue prep showed pigmented, curved conidia with 2-3 transverse septations consistent with Curvularia spp (Image 4). All blood cultures were finalized as no growth after 5 days. Fungitell was found to be greater than 500 pg/ml and Aspergillus galactomannan was negative (<0.5).
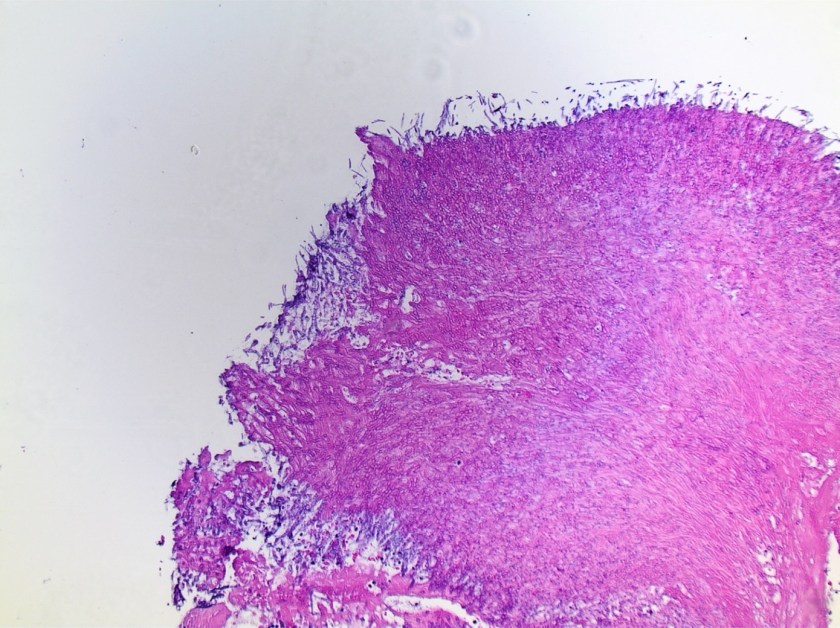
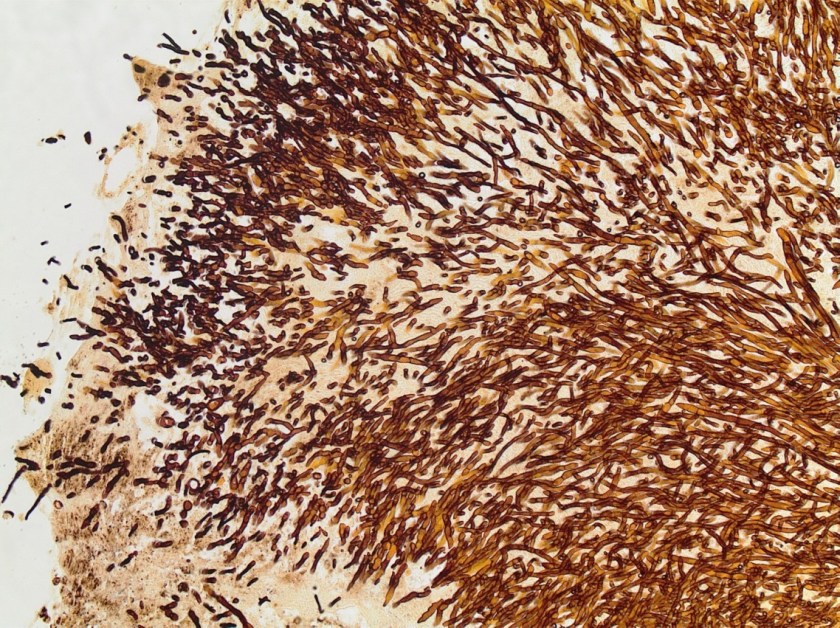

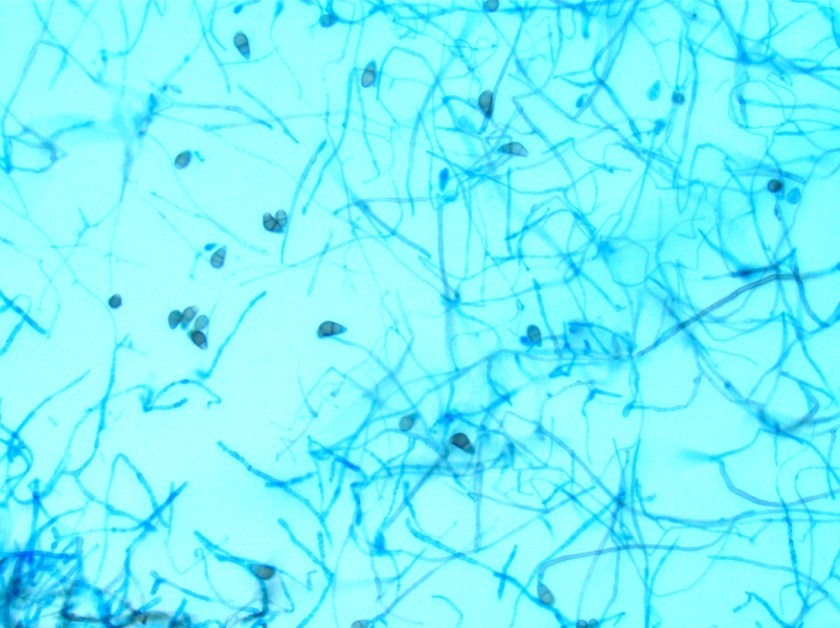
Discussion
Curvularia spp. belong to a heterogeneous group of dematiaceous or black molds. The presence of pigment in this category of molds is due to melanin in the hyphae. Dematiaceous molds are ubiquitous in nature and can occasionally cause human infections. These molds have a characteristic dark appearance on fungal media that is often dark gray, brown or black in color. In addition, when the reverse of the plate or slant is examined, the under surface is also pigmented. Based on their growth rate, the dematiaceous fungi are divided into the fast growers, such as Curvularia, Bipolaris and Alternaria spp., which are mature in 5-7 days. The second group is slow growers that take between 7-25 days to fully mature. Examples of slow growers include Phialophora, Exophila/Wangiella, Cladosporium, Fonsecaea and Rhinocladiella spp.
Most commonly, dematiaceous molds infections usually present as phaeophyphomycosis, chromoblastomycosis or mycetomas. These three entities are cutaneous or subcutaneous mycoses that are obtained by traumatic implantation but vary from one another based on clinical features and histologic features of the mold in tissues. They are most frequently cause infection in male agricultural workers in rural areas of tropical or subtropical climates. These infections are indolent in nature but can lead to significant morbidity over time, as they are difficult to treat effectively.
In addition to the above superficial infections, Curvularia spp. has also be known to cause keratitis, sinusitis and wound infections. In immunosuppressed individuals, disseminated infections with spread to the lungs and brain have been documented. Endocarditis due to Curvularia spp. is quite rare with very few cases previously reported in the literature. On those documented, Curvularia spp. infections tend to have a predilection for prosthetic heart valves or occur after cardiac surgery. Diagnosis of infective endocarditis is difficult as symptoms are indolent and blood cultures do not have a high yield. Therefore, culture of the vegetation may be the only way to make a diagnosis.
In the microbiology laboratory, Curvularia spp. will grow on routine fungal media as a darkly pigmented mold in a relatively short time. On lactophenol cotton blue prep, Curvularia spp. produce large conidia that usually contain 4 cells that are divided by transverse septations. The conidia take on a curved appearance due to swelling of the subterminal cell, which is often the largest and most deeply pigmented. If identification is necessary beyond the genus level, panfungal PCR assays followed by sequencing of ribosomal genes may be useful in providing a species level diagnosis from fresh or paraffin embedded tissue.
For localized infections, surgical treatment alone may be adequate in some cases. In infections that are extensive or if there is dissemination, treatment with newer triazoles, such as posaconazole or voriconazole, have shown a broad spectrum of activity against dematiceous molds. Amphotericin B is also another effective option. While susceptibility testing can be performed on clinically significant Curvularia spp. infections, interpretative breakpoints have not been defined and clinical correlation is lacking.
In the case of our patient, she remained on a ventilator following surgery and with the identification of mold on histology, she was started on micafungin. She was switched to amphotericin B after the mold was classified as Curvularia spp. Her condition did not improve despite therapy and she died 3 weeks after surgery.
-Azniv Azar, MD, is a fourth year anatomical and clinical pathology resident at the University of Mississippi Medical Center.

-Lisa Stempak, MD, is an Assistant Professor of Pathology at the University of Mississippi Medical Center in Jackson, MS. She is certified by the American Board of Pathology in Anatomic and Clinical Pathology as well as Medical Microbiology. She is the director of the Microbiology and Serology Laboratories. Her interests include infectious disease histology, process and quality improvement, and resident education.