A 28 year old woman underwent an elective myomectomy for menorrhagia caused by fibroids. Postoperatively, she developed fevers. A CT scan of the abdomen and pelvis showed a complex pelvic fluid collection measuring 5.4 by 4.4 by 7.0 cm. Drainage was attempted by Interventional Radiology without success. She then underwent an exploratory laparotomy with drainage of the collection, evacuation of a hematoma, and removal of an IUD. The abscess fluid was sent to the Microbiology lab for culture.
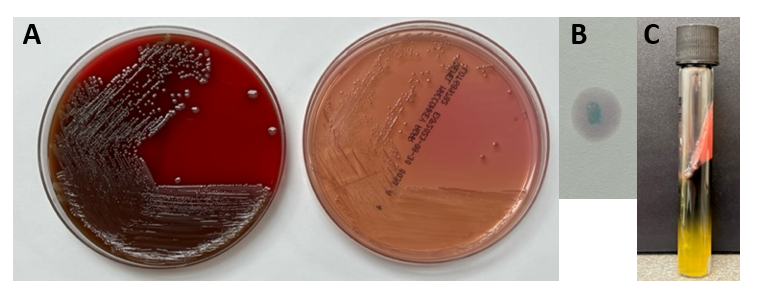
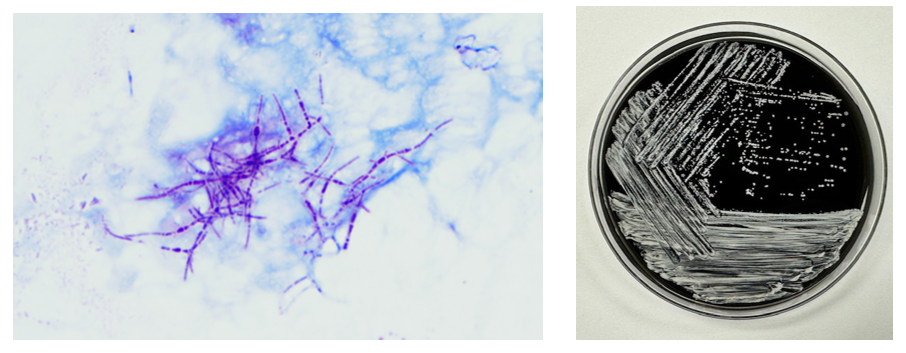
The Gram stain of the fluid revealed 1+PMN with no organisms. After 48 hours of incubation, there was few to moderate growth of pinpoint clear colonies on blood agar, with characteristic miniscule appearance of a central area of dense growth and peripherally less dense (Figure 1). Upon closeup reviewing of the colonies revealed a “fried egg” appearance (Fig 2).
There was no growth on MacConkey plate. Final identification by MALDI-TOF demonstrated Mycoplasma hominis.


Discussion
Mycoplasma hominis is a facultative anaerobe of the Mycoplasma genus, which is among the smallest free-living organisms known. They are fastidious and differentiated from other bacteria by their small size and absence of cell walls. Infections with M. hominis predominately involve the urogenital tract of females, causing pelvic inflammatory disease, bacterial vaginosis, and postpartum fever.1 Extragenital manifestations of M. hominis infections are rare but include extragenital abscesses, CNS infections in neonates, and septic arthritis.2 Reports of Mycoplasma hominis infections vary based on demographics (country, age, and number of sexual partners) but have been isolated in 4 to 30% of urogenital infections in females. 1,3
The lack of cell walls confers both diagnostic and clinical challenges. Mycoplasmas is not seen on the routine Gram stain smears. Instead of a cell wall, they have a triple-layered membrane composed of sterol.3 When able to grow in culture, colonies are small and have a ‘fried egg’ appearance on agar.4 Of the Mycoplasma species, M. hominis is the least fastidious and the most common Mycoplasma isolated on conventional culture agar media. 4-6 Non-traditional culture-based diagnosis is often made via molecular testing. 7 It is often detected in coinfections with Trichomonas vaginalis and thought to be a symbiotic relationship. 8-9
Since M. hominis is also found to be colonized in the genital tract of normal healthy individuals, it can be challenging to establish the clinical and diagnostic significance of M. hominis. With evolving molecular diagnostic assays targeting STI agents, it has been a controversial topic for assay manufacturers or labs that develop their own in-house assays whether there is a clinical utility for M. hominis PCR as part of STI multiplex PCR panels.
In contrast to Mycoplasma pneumoniae, M. hominis has intrinsic resistance to macrolides. 10 Preferred treatment regimens for M hominis infections include tetracyclines, clindamycin and fluoroquinolones. 11 However, resistance to members of these antibiotic classes has been reported and differs based on country of origin. 11 The absence of a cell wall explains the inherent resistance of Mycoplasma species to the beta-lactam antibiotic class. Its isolation in coinfections, particularly Trichomonas has been controversial in its contribution to emerging Trichomonas metronidazole resistance. 12
References
- Verteramo, R., Pastella, A., Calzolari, E., et al. An epidemiological survey of Mycoplasma hominis and Ureaplasma urealyiticum in gynaecological outpatients, Rome, Italy. Epidemiology & Infection,2013, 141(12), 2650-2657
- Zheng X, Olson DA, Tully JG, Watson HL, Cassell Gh, Gustafson DR, Svien KA, Smith TF: Isolation of Mycoplasma hominis from a brain abscess. J Clin Microbiol. 1997, 35: 992-994.
- Thomas Prescott Atkinson, Mitchell F. Balish, Ken B. Waites, Epidemiology, clinical manifestations, pathogenesis and laboratory detection of Mycoplasma pneumoniae infections, FEMS Microbiology Reviews, Volume 32, Issue 6, November 2008, Pages 956–973
- Razin S. Mycoplasmas. In: Baron S, editor. Medical Microbiology. 4th edition. Galveston (TX): University of Texas Medical Branch at Galveston; 1996. Chapter 37.
- Meloni GA, Bertoloni G, Busolo F, Conventi L. Colony morphology, ultrastructure and morphogenesis in Mycoplasma hominis, Acholeplasma laidlawii and Ureaplasma urealyticum. J Gen Microbiol. 1980;116(2):435-443.
- Christiansen G, Jensen LT, Boesen T, Emmersen J, Ladefoged SA, Schiotz LK, Birkelund S: Molecular biology of Mycoplasma. Wien Klin Wochenschr. 1997, 109: 557-561
- Baczynska, A., Svenstrup, H.F., Fedder, J. et al. Development of real-time PCR for detection of Mycoplasma hominis. BMC Microbiol 4, 35 (2004).
- Tine RC, Dia L, Sylla K, Sow D, Lelo S, Ndour CT. Trichomonas vaginalis and Mycoplasma infections among women with vaginal discharge at Fann teaching hospital in Senegal. Trop Parasitol. 2019 Jan-Jun;9(1):45-53.
- Vancini, R.G., Benchimol, M. Entry and intracellular location of Mycoplasma hominis in Trichomonas vaginalis . Arch Microbiol 189, 7–18 (2008).
- Pereyre S, Gonzalez P, De Barbeyrac B, Darnige A, Renaudin H, Charron A, Raherison S, Bébéar C, Bébéar CM. Mutations in 23S rRNA account for intrinsic resistance to macrolides in Mycoplasma hominis and Mycoplasma fermentans and for acquired resistance to macrolides in M. hominis. Antimicrob Agents Chemother. 2002 Oct;46(10):3142-50.
- Krausse R, Schubert S. In-vitro activities of tetracyclines, macrolides, fluoroquinolones and clindamycin against Mycoplasma hominis and Ureaplasma ssp. isolated in Germany over 20 years. Clin Microbiol Infect. 2010;16(11):1649-1655.
- Dessì, D., Margarita, V., Cocco, A., Marongiu, A., Fiori, P., & Rappelli, P. (2019). Trichomonas vaginalis and Mycoplasma hominis: New tales of two old friends. Parasitology, 146(9), 1150-1155. doi:10.1017/S0031182018002135

-Dr. Alex Shaffer is a first-year ID fellow (2023-2025) at Montefiore Einstein. Alex is interested in the diagnostic stewardship projects and has been actively involved in various activities with ID and micro lab faculty.

-Phyu Thwe, Ph.D, D(ABMM), MLS(ASCP)CM is Associate Director of Infectious Disease Testing Laboratory at Montefiore Medical Center, Bronx, NY. She completed her medical and public health microbiology fellowship in University of Texas Medical Branch (UTMB), Galveston, TX. Her interests includes appropriate test utilization, diagnostic stewardship, development of molecular infectious disease testing, and extrapulmonary tuberculosis.