Case description
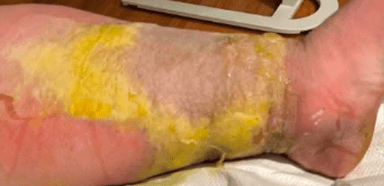
A 67 year old male presented at the clinic with a primary complaint of foot pain; she has a previous medical history of M. tuberculosis infection of her prosthetic joint, osteoarthritis, and leukopenia. The patient described joint pains during the check-up and mentioned that she also started to have periumbilical pain two weeks ago, along with worm-like objects in her stool. The patient was in Ethiopia for 8 months in the past year and was very active. He has had some weight loss but no change in appetite; he denies any diarrhea, skin rashes, fever, or chills. The patient consumed undercooked meat products during the time she visited Ethiopia. No abnormal neurological symptoms presented at the time of the visit.
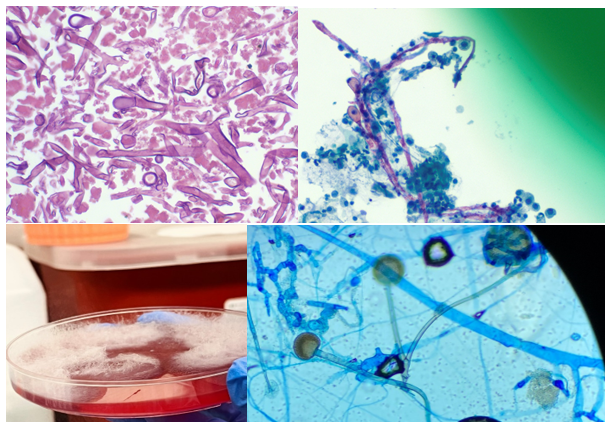
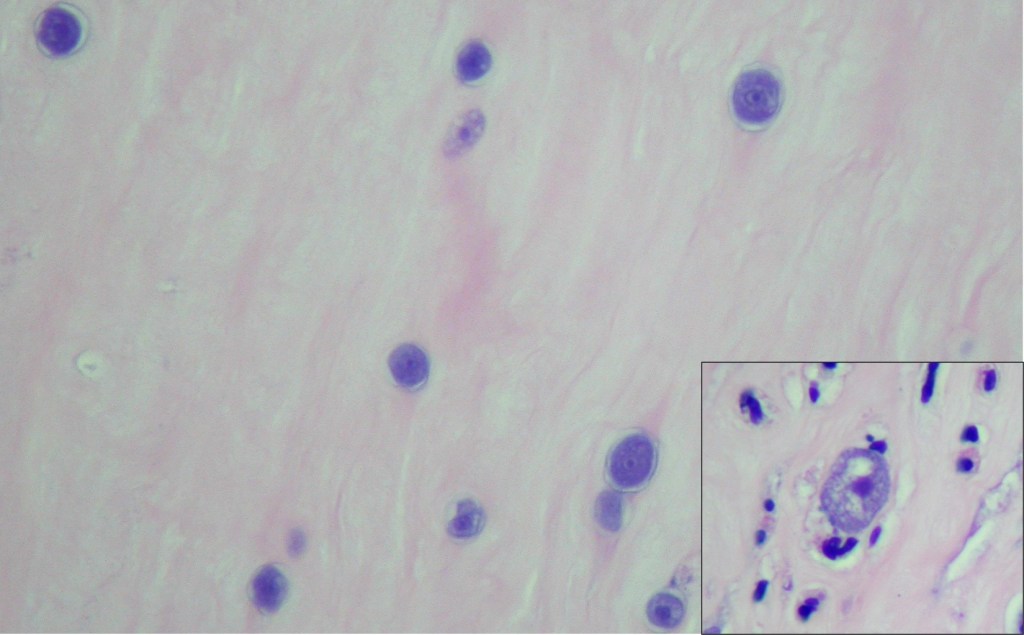
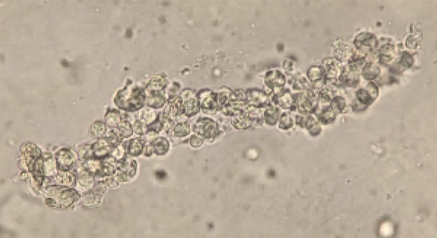
Orders were placed for H. Pylori antigen, fecal bacteria pathogen PCR, Giardia and Cryptosporidium antigen, and Ova & Parasite exam for the patient’s GI symptoms. The Ova & Parasite exam detected the objects in Image 1.

Discussion
The Ova & Parasite exam was reported as Taenia species. The eggs had a diameter of around 37um. An infectious disease consult was ordered and a single dose of 600mg praziquantel was prescribed for the treatment. Repeat Ova & Parasite exams are ordered for 3 days post-treatment looking for dying parasites and 1 month post-treatment to confirm the cure (no eggs).
Taenia in the Taeniidae family of tapeworms (BioLib, n.d.). Three species are commonly found and most clinically important in human infection: Taenia saginata, Taenia solium, and Taenia asiatica; most Taeniasis is asymptomatic or has mild symptoms (Centers for, 2020b).
Taenia solium, or pork tapeworm often found in pork, is the most dangerous species to humans for two reasons. First, this is the only species that can cause the neurologic symptoms by cysticercosis in brain tissue; second, this species can take humans as intermediate hosts, which means it can cause human to human transmission within the household (Schmidt et al., 2009).
Taenia asiatica also lives in pigs, primarily in the liver instead of muscle. This species has a very similar genetic, morphology, and immunology to T. saginata. It is frequently found in Asia (Schmidt et al., 2009).
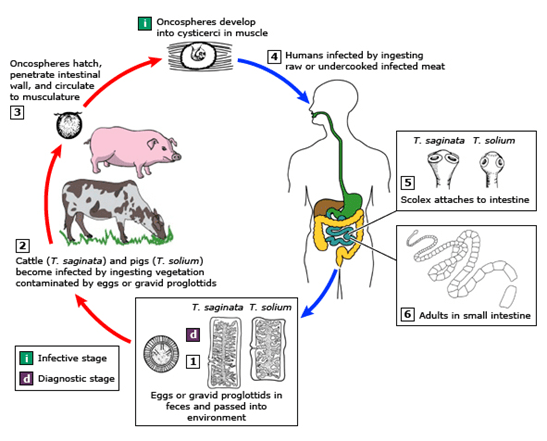
Taenia saginata, or beef tapeworm, is what our patient was assumed to have in this case. The life cycle is shown below in Figure 2. The patient presented because his ankle pain started to impact his walking significantly; however, he was not seeking help for his worm-like objects in the clinic, probably due to the mildness of the symptoms. The parasite infection was brought into sight because of his travel history and stool observation. Per CDC, Eastern Europe, Russia, eastern Africa, and Latin America are the highest risk areas (Centers for, 2020a). The patient stayed for 8 months in Ethiopia in eastern Africa. Ethiopia has a relatively poor sanitation status and a high prevalence of taeniasis (Jorga, 2020). The major contributors for our infectious disease clinicians to assume this patient has T. saginata infection but not T. solium infection are: there are no neurological symptoms, and there is no pork exposure due to his religion. Visualization of the tapeworm eggs or segments is important for identification the species. In this case, many eggs were found on the wet mount slide from the patient’s stool sample.
Treatment of taeniasis is with Praziquantel. Praziquantel removes the tapeworms from the human body by detaching the worm suckers from vessel walls. The medication is safe to give to ≥1year old patients (UpToDate, 2022).
https://www.uptodate.com/contents/image/print?imageKey=ID%2F64879
References
BioLib: Biological library. Taenia | BioLib.cz. (n.d.). Retrieved from https://www.biolib.cz/en/taxon/id43806/
Centers for Disease Control and Prevention. (2020a, September 18). CDC – taeniasis – general information . Epidemiology & Risk Factors. Retrieved from https://www.cdc.gov/parasites/taeniasis/epi.html
Centers for Disease Control and Prevention. (2020b, September 18). CDC – taeniasis – general information . frequently asked questions. Retrieved from https://www.cdc.gov/parasites/taeniasis/gen_info/faqs.html
Jorga, E., Van Damme, I., Mideksa, B. et al. Identification of risk areas and practices for Taenia saginata taeniosis/cysticercosis in Ethiopia: a systematic review and meta-analysis. Parasites Vectors 13, 375 (2020). https://doi.org/10.1186/s13071-020-04222-y
Schmidt, G. D., & Roberts, L. S. (2009). Chapter 21 Tapeworms. In Foundations of Parasitology, eighth edition (pp. 346–351). essay, McGraw-Hill Higher Education.
UpToDate. (2022). Praziquantel: Drug information. UpToDate. Retrieved from https://www.uptodate.com/contents/table-of-contents/drug-information
-Sherry Xu is a Masters student in the department of Pathology and Laboratory Medicine at the University of Vermont Medical Center.

-Christi Wojewoda, MD, is the Director of Clinical Microbiology at the University of Vermont Medical Center and an Associate Professor at the University of Vermont.