Much has changed quickly with SARS-CoV-2 virus (COVID-19) testing. Several commercial options are now available. Labs have less problems getting control material (positive samples are no longer in short supply). And labs that opted to bring on testing are now running multiple versions of COVID-19 molecular tests with a combination of high speed platforms or high throughput. Rapid cartridge tests are used for clearing people from the ED/ removing contact isolation on inpatients while the high throughput assays are used for routine screening.
However, several bottlenecks still exist. There are shortages of nucleic acid extraction kits, collection swabs and viral transport media. Fortunately, some recent studies have demonstrated preliminary evidence for using alternative sample types, collection methods, and storage conditions.
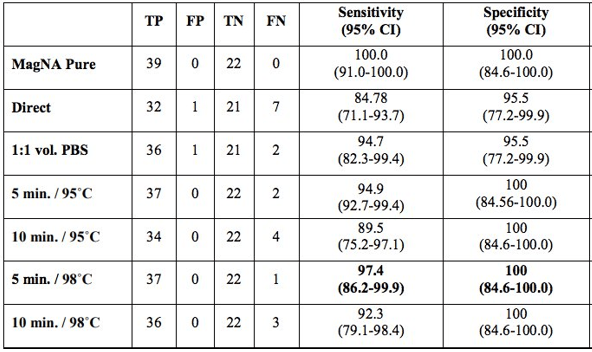
One of the first tenets of molecular diagnostics is isolation and purification of nucleic acid. Therefore, it was surprising to see a report on an extraction-free COVID-19 protocol from Vermont (Bruce EA et al.). This study initially analyzed two patient samples and showed drops in sensitivity of ~4Ct cycles. While this would not be suitable for low level detection, many viral samples have high levels of virus that still would permit detection. The team went on to test this method on 150 positive specimens from the University of Washington and found 92% sensitivity with 35% sensitivity at the low viral load range (Ct value> 30). This was improved with a brief heat inactivation step (Table 1). This was similarly seen in a study from Denmark, where brief heat inactivation of extraction-free methods (Direct) had 97% specificity in 87 specimens (Table 2).

Some similar studies out of Chile also showed extraction-free protocols on a larger number of specimens, and they reported a loss in sensitivity varying from 1-7 Ct cycles depending on the primers used.

As this novel Coronavirus has an RNA-based genome, RNA is the target of molecular tests. As RNA is susceptible to degradation, there have been concerns over sample storage. Should it be refrigerated? Frozen? How do multiple freeze-thaw cycles impact specimen stability? Are there viable alternatives to viral transport media? One preliminary study explored these questions very nicely. They took X multiple sample types (NP, BAL, saline storage media) and stored them at 20C, 4C, -20, and -70 for multiple days up to 1 week and then analyzed the level of virus detected. In each case, the loss in sensitivity was minimal (<2 Ct cycles from day 0 to day 7) at room temperature with comparable results at lower temperatures (Table 3).

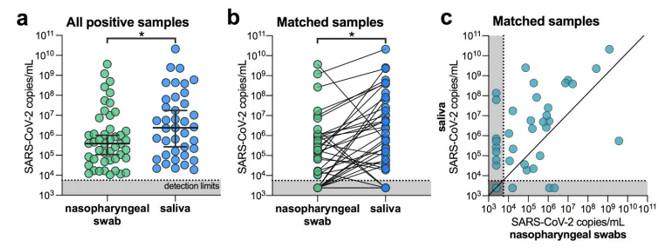
Lastly, alternative sample types such as saliva will help break the bottleneck in swabs and viral transport media. I was surprised to hear about this being a suitable alternative. Having worked with saliva for DNA analysis, I know it can be contaminated, of variable quantity, includes digestive enzymes and is viscous (slimy). These are not characteristics a lab would look for in a specimen type being used for high-throughput testing where several sample failures could occur. But these researchers from Yale showed measurable levels of SARS-CoV-2 that facilitated even higher sensitivity than nasopharyngeal swabs (Wylie AL et al).
With a much-needed increase in testing for this country, optimizations need to be implemented to improve efficiency. These steps alone will not be enough, but if we can have extraction-free testing of saliva collected at home, this would provide a substantial benefit to bringing easy testing to everyone.
UPDATE: Since this was written, the first FDA EUA was authorized for an at-home saliva collection kit for use at the Rutger’s clinical genomics lab (https://www.fda.gov/media/137773/download).
References
Please note: many of these references were on pre-print servers and have not been peer-reviewed.
- Bruce EA, Huang ML, Perchetti GA, et al. DIRECT RT-qPCR DETECTION OF SARS-CoV-2 RNA FROM PATIENT NASOPHARYNGEAL SWABS WITHOUT AN RNA EXTRACTION STEP. 2020. https://www.biorxiv.org/content/10.1101/2020.03.20.001008v2.full#T2
- Wyllie AL, Fournier J, Casanovas-Massana A, Campbell M et al. Saliva is more sensitive for SARS-CoV-2 detection in COVID-19 patients than nasopharyngeal swabs. medRxiv 2020. https://www.medrxiv.org/content/10.1101/2020.04.16.20067835v1#disqus_thread
- Fomsgaard AS, Rosentierne MW. An alternative workflow for molecular detection of SARS-CoV-2 – escape from the NA extraction kit-shortage, Copenhagen, Denmark, March 2020. https://www.medrxiv.org/content/10.1101/2020.03.27.20044495v1.full.pdf
- Rogers AA, Baumann RE, Borillo GA, et al. Evaluation of Transport Media and Specimen Transport Conditions for the Detection of SARS-CoV-2 2 Using Real Time Reverse Transcription PCR. JCM 2020.
- Beltran-Pavez C, Marquez CL, Munoz G et al. SARS-CoV-2 detection from nasopharyngeal swab samples without RNA extraction. bioRxiv 2020. https://www.biorxiv.org/content/10.1101/2020.03.28.013508v1.full.pdf

-Jeff SoRelle, MD is a Chief Resident of Pathology at the University of Texas Southwestern Medical Center in Dallas, TX. His clinical research interests include understanding how the lab intersects with transgender healthcare and improving genetic variant interpretation.
