Methicillin-resistant Staphylococcus aureus (MRSA) is a well-known cause of bacteremia, pneumonia, skin and soft tissue infections, and osteomyelitis, resulting in significant morbidity and mortality worldwide.1 Many testing methods (e.g. MALDI-TOF with susceptibility testing, molecular, chromogenic agar) have been developed for identification of MRSA and clinical microbiology laboratories will often use more than one. On occasion this leads to discrepant results which can be challenging to resolve and report.
How does methicillin resistance work?
Staphylococcus aureus (SA)has a peptidoglycan cell wall containing alternating N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM) molecules with peptide chains reinforced by crosslinks. Crosslinking is mediated by penicillin-binding proteins (PBPs), which are the targets of beta-lactam antibiotics such as penicillins and cephalosporins.2 In methicillin-sensitive S. aureus (MSSA), these antibiotics bind PBPs and prevent formation of crosslinks, thus disrupting cell wall synthesis. However, methicillin resistance can occur if the PBPs are altered. MRSA produces PBP homologues such as PBP2a (encoded by the mecA gene) or more rarely, PBP2c (encoded by mecC), which don’t allow beta-lactam antibiotics to bind strongly so crosslinking occurs.3,4
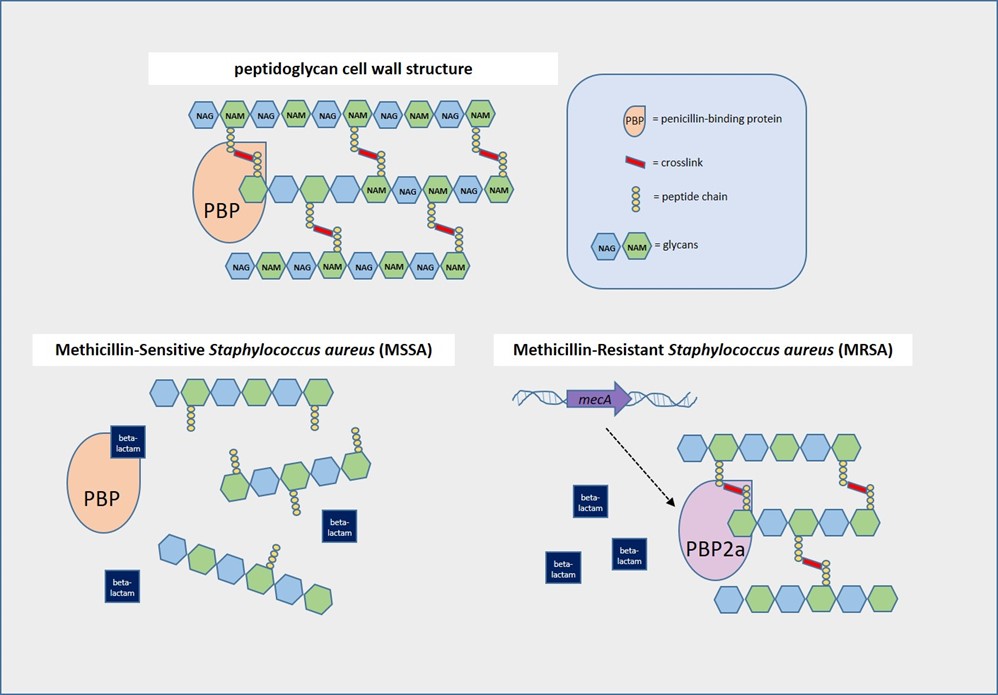
What tests are used to identify MRSA?
MRSA testing can be genotypic or phenotypic, but most cannot be performed directly on patient samples. With molecular testing, we can detect mecA and/or mecC, the genes most commonly responsible for methicillin resistance. However, positive molecular results on a direct specimen source (e.g., positive blood culture) cannot be definitively attributed to SAif other mecA-harboring organisms such as methicillin-resistant Staphylococcus epidermidis are also present.5
When there is a pure isolate of SA growing in culture, lateral flow assays and latex agglutination tests can be used to interrogate the presence of mecA. Both lateral flow assays and latex agglutination tests detect PBP2a using antibodies specific to this alternative penicillin-binding protein. Chromogenic agars are a modern-day biochemical test, taking advantage of specific enzymes produced by MRSA (e.g. phosphatase) which cleave chromogens in the media.6
Disk diffusion and broth/agar dilution are the standard phenotypic methods for quantitating antimicrobial resistance in SA growing in bacterial culture. Despite the name, methicillin is no longer used for testing or treatment of MRSA. Per Clinical and Laboratory Standards Institute, oxacillin-resistant and cefoxitin-resistant SA should both be reported as MRSA and considered resistant to all beta-lactam antibiotics.7
Why don’t my test results match?
Although detection of the mecA gene or its protein product PBP2a are the standard7, mixed MSSA and MRSA cultures can lead to discrepant results. Another source of genotypic-phenotypic discrepancy are mecA mutations where the gene is still present and detected, but functional PBP2a is no longer produced. PBP2c only shares ~70% homology to PBP2aand is not detected by latex agglutination assays4-5, and mecC-mediated MRSA might be resistant only to cefoxitin and not oxacillin7. Other mechanisms of MRSA resistance are still being studied and not all are included on molecular test panels.
References
- Turner, N.A., Sharma-Kuinkel, B.K., Maskarinec, S.A. et al. Methicillin-resistant Staphylococcus aureus: an overview of basic and clinical research. Nat Rev Microbiol 17, 203–218 (2019). https://doi.org/10.1038/s41579-018-0147-4
- Sawa, T., Kooguchi, K. & Moriyama, K. Molecular diversity of extended-spectrum β-lactamases and carbapenemases, and antimicrobial resistance. j intensive care 8, 13 (2020). https://doi.org/10.1186/s40560-020-0429-6
- Srisuknimit V, Qiao Y, Schaefer K, Kahne D, Walker S. Peptidoglycan Cross-Linking Preferences of Staphylococcus aureus Penicillin-Binding Proteins Have Implications for Treating MRSA Infections. J Am Chem Soc. 2017 Jul 26;139(29):9791-9794. doi: 10.1021/jacs.7b04881.
- Ballhausen B, Kriegeskorte A, Schleimer N, Peters G, Becker K. The mecA homolog mecC confers resistance against β-lactams in Staphylococcus aureus irrespective of the genetic strain background. Antimicrob Agents Chemother. 2014 Jul;58(7):3791-8. doi: 10.1128/AAC.02731-13.
- Lakhundi S, Zhang K. Methicillin-Resistant Staphylococcus aureus: Molecular Characterization, Evolution, and Epidemiology. Clin Microbiol Rev. 2018 Sep 12;31(4):e00020-18. doi: 10.1128/CMR.00020-18.
- Flayhart D, Hindler JF, Bruckner DA, et al. Multicenter evaluation of BBL CHROMagar MRSA medium for direct detection of methicillin-resistant Staphylococcus aureus from surveillance cultures of the anterior nares. J Clin Microbiol. 2005;43(11):5536-5540. doi:10.1128/JCM.43.11.5536-5540.2005
- CLSI Performance Standards for Antimicrobial Susceptibility Testing M100, 32nd edition. (2022) Clinical and Laboratory Standards Institute

– Angelica Moran, MD, PhD is a clinical microbiology fellow at University of Chicago Medicine and NorthShore University Healthsystem and research fellow at the Duchossois Family Institute. She is interested in translational research developing clinical laboratory diagnostics for precision medicine and the microbiome.

-Paige M.K. Larkin, PhD, D(ABMM), M(ASCP)CM is the Director of Molecular Microbiology and Associate Director of Clinical Microbiology at NorthShore University HealthSystem in Evanston, IL. Her interests include mycology, mycobacteriology, point-of-care testing, and molecular diagnostics, especially next generation sequencing.
